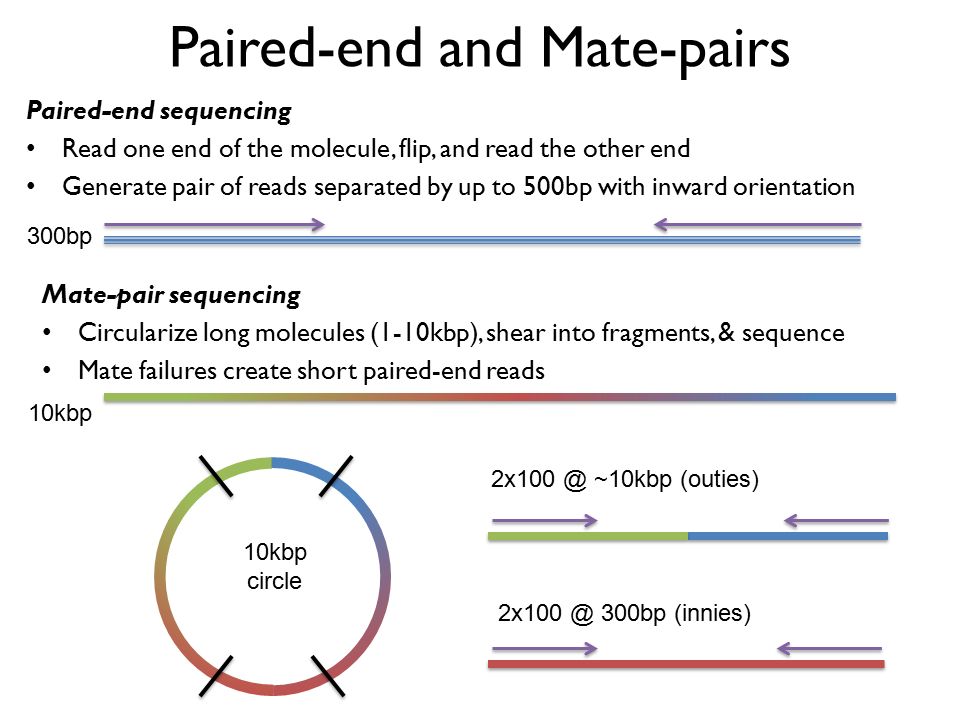
paired end sequencing vs mate pair
Paired end や mate pair という用語はどのようにライブラリが作られたかどうやってシーケンスされたかを示します. In addition to producing twice the number of reads for the same time and effort in library.

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology
Usually mate-pair library are used to identify structural.
. Standard libraries depends upon your application. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. To simplify you can differ between two kinds of reads for paired-end sequencing.
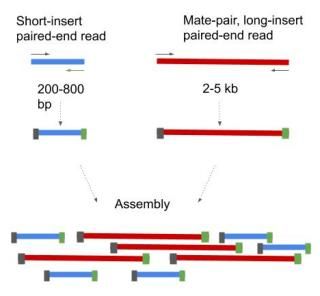
Mate pairs is an obsolete type of sequencing library method for obtaining long distance information. The difference between the two variants is first surprise - the length of the insert. SIPERs are 200800 bp long LIPERs can be longer.
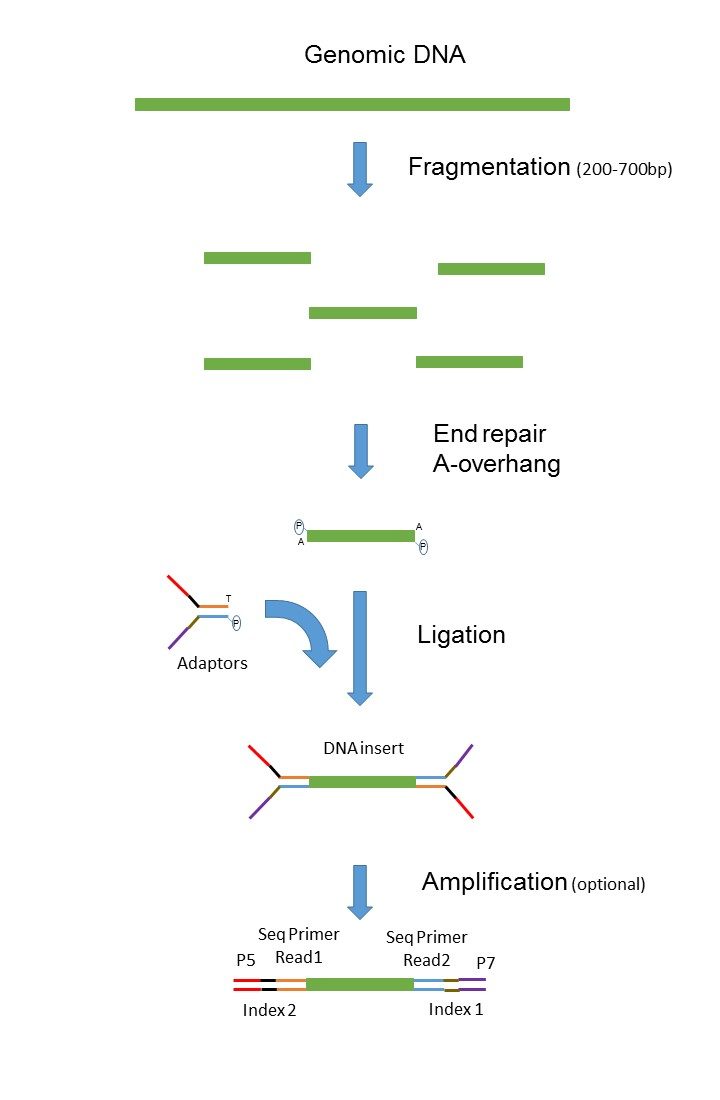
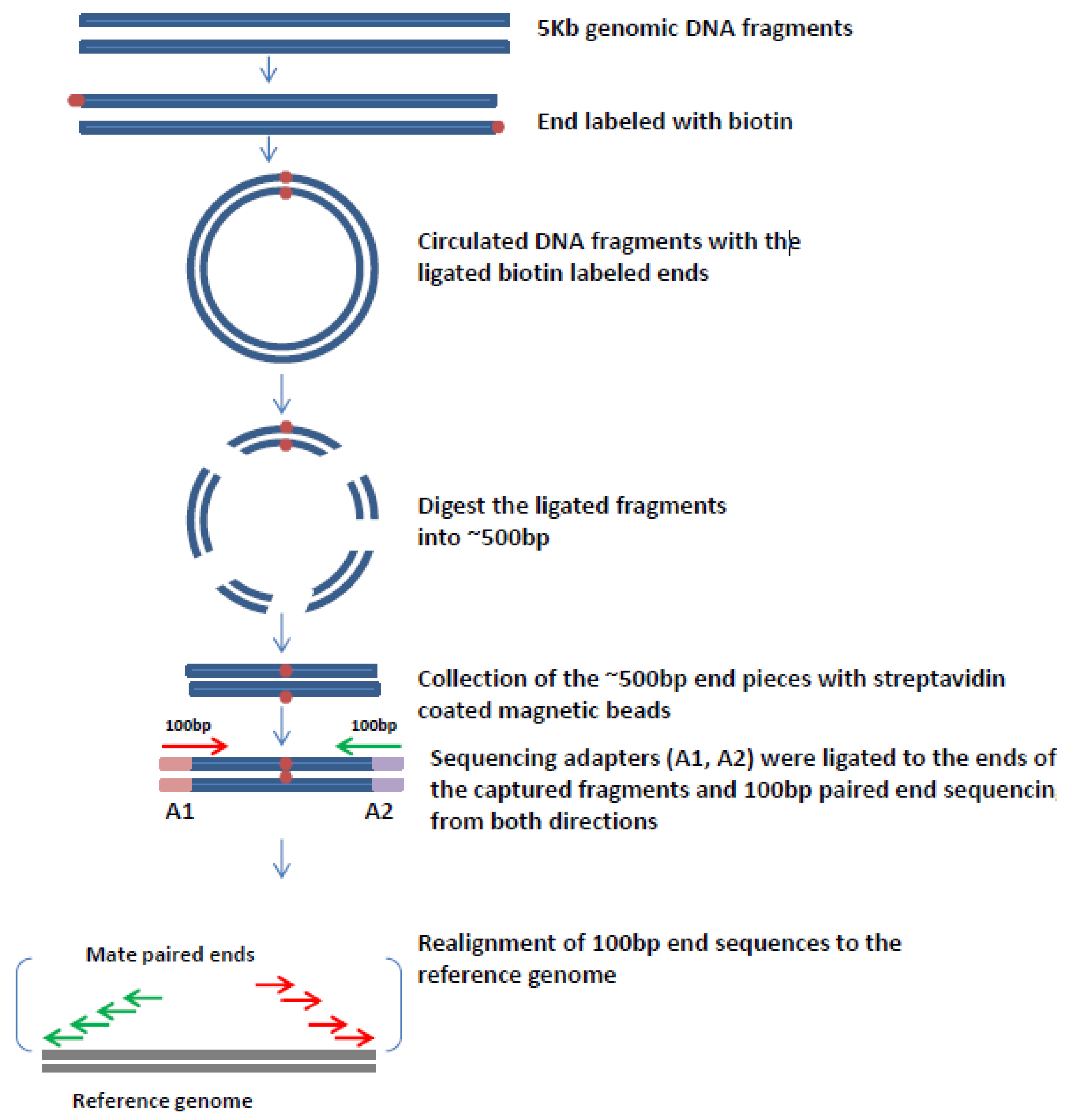
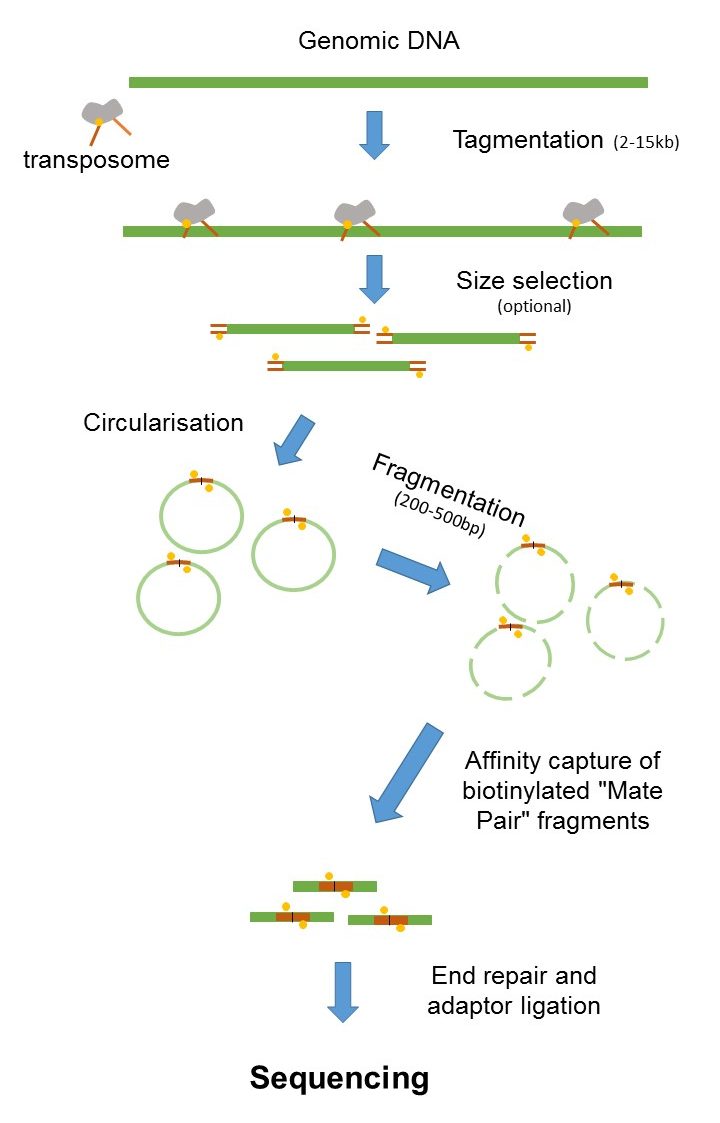
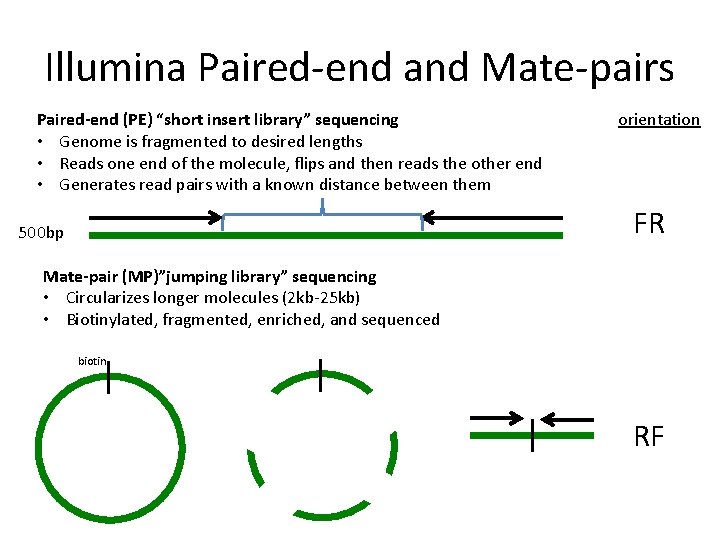
While the insert size of mate. In mate-pair libraries you are creating a library of fragment ends and then performing normal sequencing on these end fragments. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. So if you have a 500bp fragment you machine will sequence 1-75 and 425-500 of the fragment. They differ in library preparation.
Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. The decision to use mate-pair vs. Paired-end is a type of sequencing.
In short-read sequencing intact genomic DNA is sheared into several million short DNA fragments called reads. Shortinsert pairedend reads SIPERs and long-insert paired-end reads LIPERs. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
In pair-end sequencing the machine is sequencing the ends of a fragment. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall. The similarities between PE and MP reads include.
Individual reads can be paired together to create paired-end reads which offers some benefits for downstream bioinformatics data analysis algorithms. Reads come in pairs. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
The structure of a paired-end read is described here. 例えばゲノム DNA を. In mate-pair sequencing the library preparation yields two.
I wondered what the essential differences were between paired-end mate-pair and long read. However the pair reads typically overlap for all four libraries. In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably.
In fact mate-pair libraries require paired-end sequencing. For me mate-pair and classic paired-end are both paired-end reads with the difference that. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends.
The insert size on classic paired-end is smaller about 500bp. In the range of kb ie 1kb 2kb or longer in some cases. Also these libraries have insert sizes much longer than the paired end.
Its a fun intellectual exercise but realistically it is better to delve into long reads linked reads or HiC Mate pairs derive from jumping libraries which were important pieces of mol. While the underlying principles between PE and MP reads have strong similarities there are inherent differences that are crucial to understand. Introduction to Mate Pair Sequencing.
Here is some extracted data mapping paired reads. Bases 1-75 forward direction and bases 225-300 reverse direction of the fragment. In addition to producing twice the number of reads for the same time and effort in library.
They are not two different methods. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. For example if you have a 300bp contiguous fragment the machine will sequence eg.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. I writing here because I have some questions for you. By magic-blasting the SRA paired reads against the public available assembly I expected that paired end reads would map near each other on a contigscaffold while mate pair reads would map further away on a contigscaffold.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal. Since paired-end reads are more likely to align to a reference the quality of the entire data set.
Introduction to Mate Pair Sequencing. Mate-pair is a specific type of library. The latter one is also called mate pair.
Whole Genome Assembly With Iplant Ppt Video Online Download
Paired End Sequencing France Genomique
Viruses Free Full Text Mate Pair Sequencing As A Powerful Clinical Tool For The Characterization Of Cancers With A Dna Viral Etiology Html

Ion Torrent Mate Paired Library Preparation Protocol And Sequencing Download Scientific Diagram
Mate Pair Sequencing France Genomique

Schematic Representation Of Mate Pair Library Construction By Coupling Download Scientific Diagram

Pdf Data Processing Of Nextera Mate Pair Reads On Illumina Sequencing Platforms Semantic Scholar
Why Mate Paired End Paired End And Single End Reads Library To Be Combined For Assembling

Principles For Construction Of Mate Pair Sequencing Libraries A Download Scientific Diagram

Construction Of Mate Pair Libraries Download Scientific Diagram

What Is Mate Pair Sequencing For

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download Scientific Diagram

What Is Mate Pair Sequencing For

Mate Pair Made Possible Enseqlopedia
Introduction To Genome Assembly Tutorial
De Novo Genome Assembly Using Next Generation Sequence

6 Pairwise Strand Of Paired End And Mate Pair Read Libraries Download Scientific Diagram
